Ryann Perez
Computational Biochemist
Specializing in machine learning, protein science, and generative AI. Building impactful systems at the intersection of deep learning and biology.
Computational Projects
TAsk
A RAG-based research assistant that helps reason through advanced concepts by searching through class documents and references, then generating informed responses with source citations. The capabilities and educational benefits of this system were studied in a real biological chemistry classroom.
AggBERT: Amyloid Prediction
A deep learning framework for predicting amyloid-forming hexapeptides using semi-supervised ProtBERT models. Trained on WALTZ-DB dataset with predictions across a 64M peptide manifold. A useful tool for biologic design.
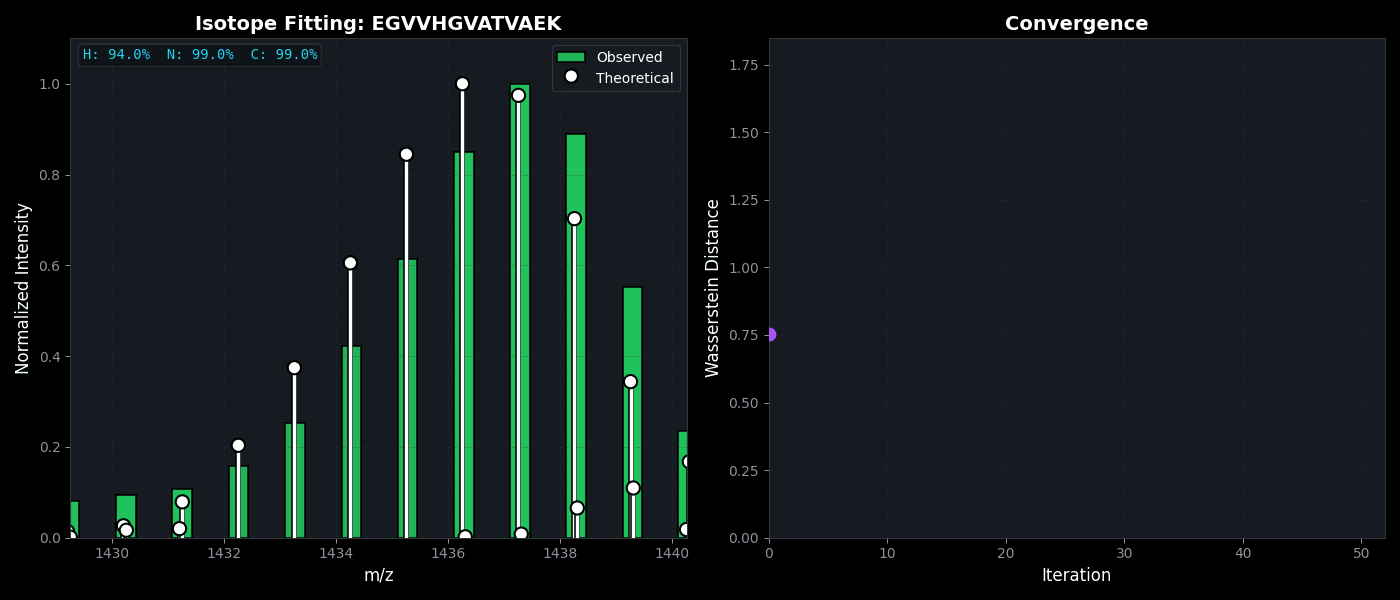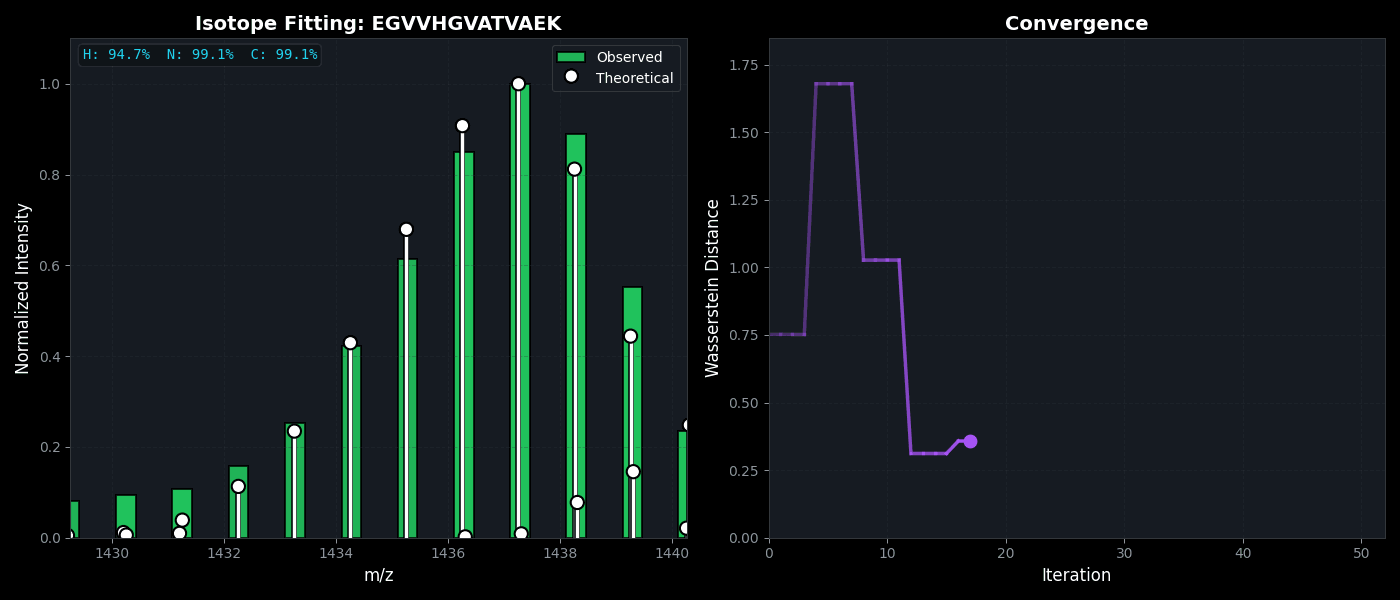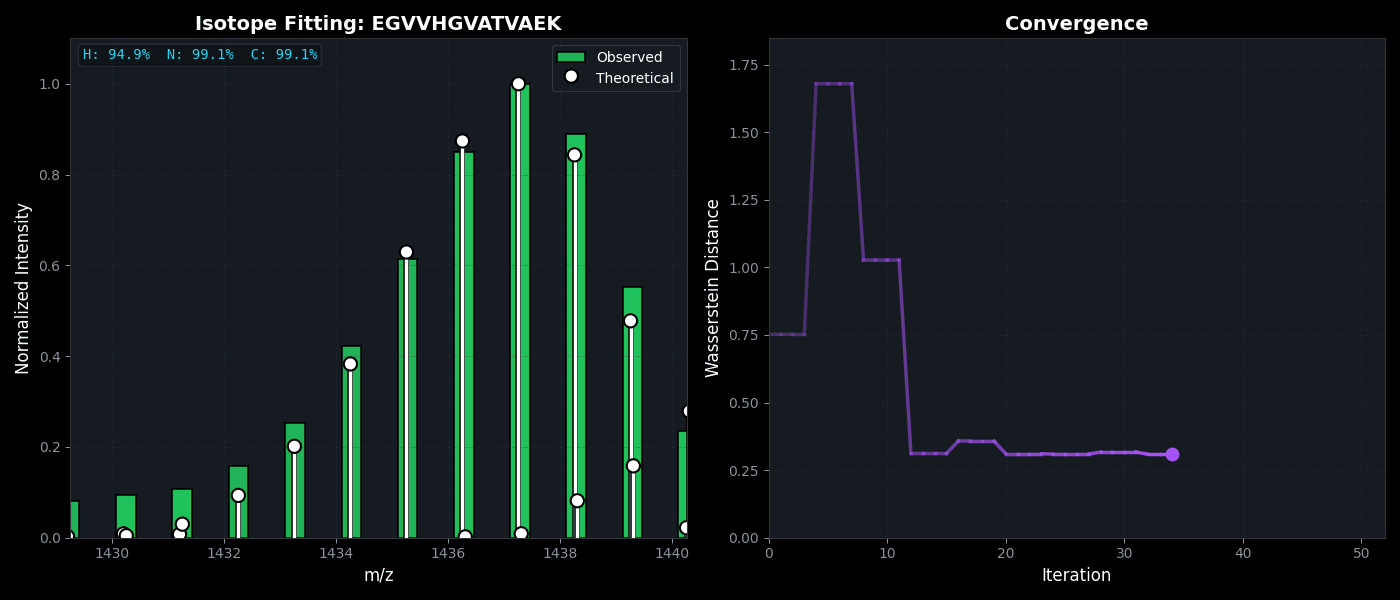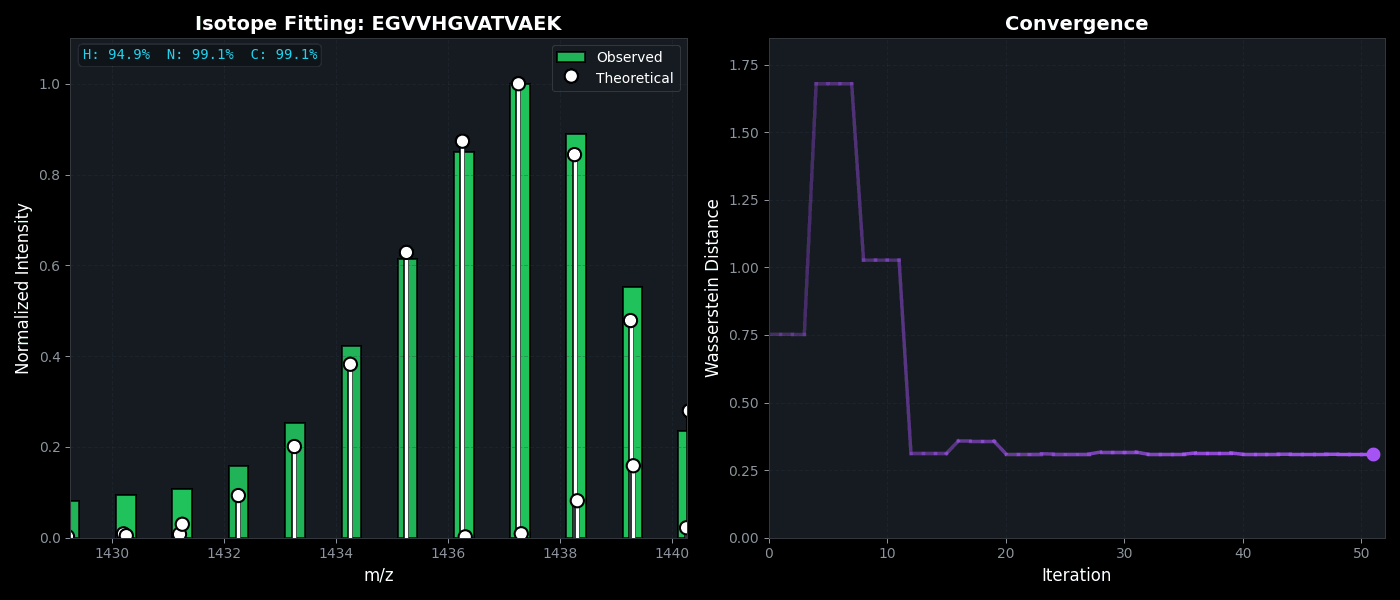
Isotope Distribution Estimation
Tools for calculating and visualizing fine isotope patterns in MALDI-TOF data. Includes methods for estimating heavy isotope incorporation fractions in tryptic peptides containing heavy C, N, or H.
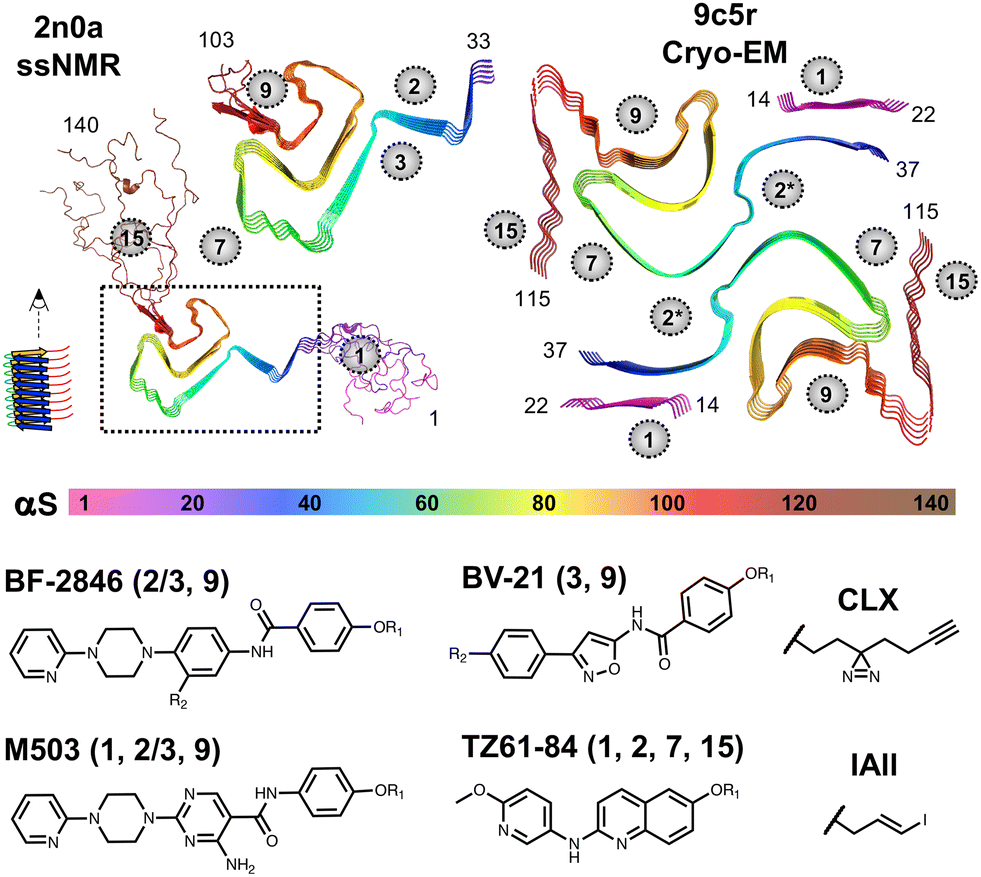
Alpha-Synuclein Binder
A machine learning framework to predict new high-affinity ligands that bind to α-synuclein fibrils, a key pathological feature of Parkinson's disease and related synucleinopathies. Trained on fewer than 300 experimentally measured binding affinities, the model identified five new sub-10 nM binders from a 140 million-compound virtual library.
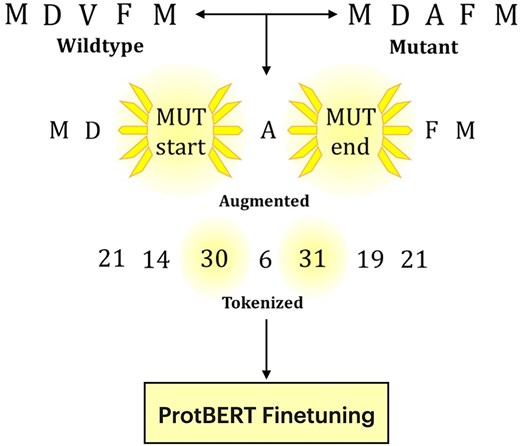
HintToken Learning
Novel approach to data augmentation for protein language models using hint tokens for improved model training and inference.
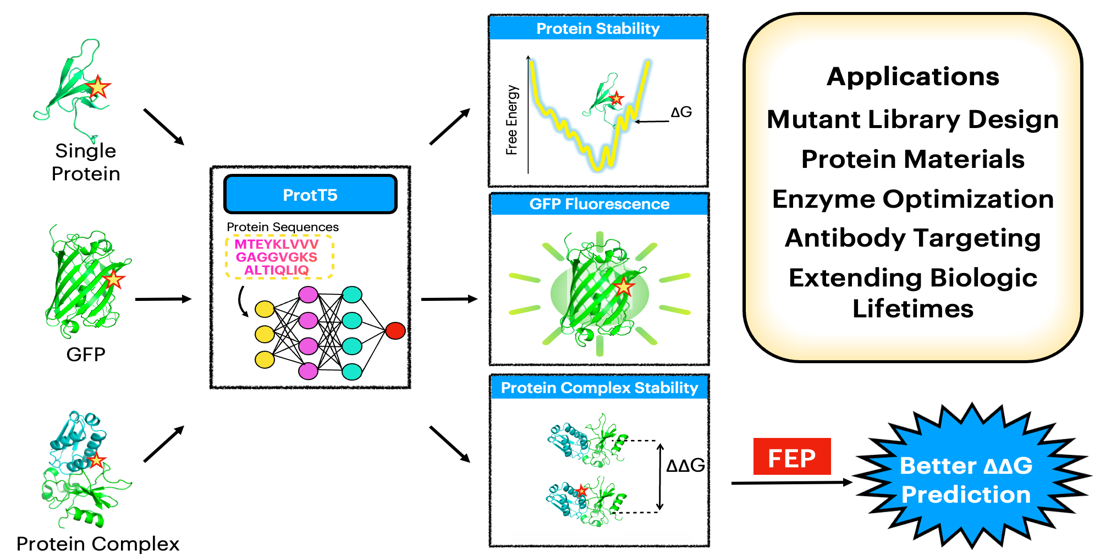
Protein Stability Prediction
Large language models for predicting protein stability changes upon mutation. Useful for protein engineering and understanding disease-causing variants.